High-Performance Python – Distributed Python

Scale Python GPU code to distributed systems and your laptop.
As the last few articles have pointed out, Python is becoming an important language in HPC and is arguably the most important language in data science. The previous three articles have covered tools for improving the performance of Python with C, Fortran, and GPUs on a single node.
In this article, I present tools for scaling Python GPU code to distributed systems. Some of the tools are Python variations of classic HPC tools, such as MPI for Python (mpi4py), a Python binding to the Message Passing Interface (MPI). Tools such as Dask focus on keeping code Pythonic, and other tools support the best performance possible.
MPI for Python
The HPC world has been using MPI since the early 1990s. In the beginning, it was used almost exclusively with C/C++ and Fortran. Over time, versions of MPI or MPI bindings were released for other languages such as Java, C#, Matlab, OCaml, R, and, of course, Python. MPI was used because it is the standard in the HPC world for exchanging data between processes on shared or distributed architectures. Python is arguably the number one language for Deep Learning and is very popular in Machine Learning and HPC. Combining the two seemed like a natural step toward improving Python performance, while making code writing for science and engineering similar to the current language and tools already in use.
MPI for Python (mpi4py) was developed for Python by using the C++ bindings on the MPI-2 standard. It has been designed to be fairly Pythonic, so if you know Python and understand the basic concepts of MPI, mpi4py shouldn't be a problem. I am a self-admitted Fortran coder and using mpi4py, even though it is based on the C++ MPI bindings, is no more difficult than with Fortran. In fact, I think it's a bit easier, because Python has data structures that can be used to create complex data structures and objects. To pass these structures between MPI ranks, Python serializes the data structures using pickle, with both ASCII and binary formats supported. Python can also use the marshal module to serialize built-in objects into a binary format specific to Python.
Mpi4py takes advantage of Python features to maximize the performance of serializing and de-serializing objects as part of data transmission. These efforts have been taken to the point that the mpi4py documentation claims these features enable "… the implementation of many algorithms involving multidimensional numeric arrays (e.g., image processing, Fast Fourier Transforms, finite difference schemes on structured Cartesian grids) directly in Python, with negligible overhead, and almost as fast as compiled Fortran, C, or C++ codes."
Mpi4py has all of the communications and functions that you expect from MPI:
- Blocking and non-blocking send/receive
- Collective communications
- Scatter/gather
- Dynamic process management (MPI-2 standard)
- One-sided communications
- Parallel input/output: (MPI.File class)
- Initialization
- Timers
- Error handling
All of these are accomplished very Pythonically but, at the heart, are still MPI.
mpi4py Example
Running mpi4py code is about the same as running classic C/Fortran code with MPI. For example, using Open MPI, the command for running MPI code would be,
$ mpiexec -n 4 python script.py
where script.py is the Python code. Depending on how Python is installed or built on your system, you might have to define the fully qualified path to the Python executable itself, the script, or both. Here is a quick example of a Hello World mpi4py code.
from __future__ import print_function
from mpi4py import MPI
comm = MPI.COMM_WORLD
print("Hello! I'm rank %d from %d running in total..." % (comm.rank, comm.size))
comm.Barrier() # wait for everybody to synchronize _here_It looks like a classic MPI program that has been "pythonized." A second example from an mpi4py tutorial illustrates how to communicative NumPy arrays using MPI:
from mpi4py import MPI import numpy comm = MPI.COMM_WORLD rank = comm.Get_rank() # passing MPI datatypes explicitly if rank == 0: data = numpy.arange(1000, dtype='i') comm.Send([data, MPI.INT], dest=1, tag=77) elif rank == 1: data = numpy.empty(1000, dtype='i') comm.Recv([data, MPI.INT], source=0, tag=77) # automatic MPI datatype discovery if rank == 0: data = numpy.arange(100, dtype=numpy.float64) comm.Send(data, dest=1, tag=13) elif rank == 1: data = numpy.empty(100, dtype=numpy.float64) comm.Recv(data, source=0, tag=13)
Don't forget that mpi4py will serialize the data when passing it to other ranks, but the good news is that it works very hard to make this as efficient as possible. The third and last example is a simple broadcast (bcast) of a Python dictionary:
from mpi4py import MPI
comm = MPI.COMM_WORLD
rank = comm.Get_rank()
if rank == 0:
data = {'key1' : [7, 2.72, 2+3j], 'key2' : ( 'abc', 'xyz')}
else:
data = None
#endif
data = comm.bcast(data, root=0)I threw in my Fortran habit of putting in a comment to mark the end of an if/then/else statement or loop (sometimes the indentation gets a little crazy).
UCX
HPC developers, which can include MPI developers and HPC system vendors, seem to create communication frameworks (communication middleware) for new concepts, which limits interoperability and means users have to "port" their applications to the frameworks if they switch software stacks. To reduce this problem, Unified Communication X (UCX) was created by a multivendor consortium. UCX is an "… open-source, product grade, communication framework focusing on data-centric and high performance applications." The goal of UCX is to provide the functionality needed for efficient high-performance communication, eliminating the need to "roll your own" and reducing the porting time when different software stacks are involved.
UCX has a set of interfaces to various libraries that support networks such as InfiniBand and NVLink. Many tools can be built on top of UCX taking advantage of the commonality of UCX as well as the performance and portability. For example, UCX is currently a framework used within Open MPI. You can also create bindings on top of UCX to provide fast, low-latency data communications for various networks. UCX has three parts to its framework: (1) UC-P for Protocols, (2) UC-T for Transport, and (3) UC-S for Services. UC-P is a high-level API that uses the UCT transport layer framework to construct common protocols and is useful for things such as multirail support, device selection, tag-matching, and so on. UC-T for Transport is a low-level API that exposes basic network operations used by the network hardware. Finally, UC-S for Services provides the infrastructure for component-based programming, memory management, and other services and is primarily used for platform abstractions, data structures, and so on. These APIs are used within UCX to achieve the goals of portability, high bandwidth, and low latency.
UCX Python (UCX-Py)
The RAPIDS project develops Machine Learning tools and algorithms on top of GPUs, which is needed for high-speed networking for Dask (discussed in next section). Moreover, it needs a communication framework that can take advantage of GPU networking (NVLink). Because UCX provided these features, UCX Python, also referred to as UCX-Py, was created. Like RAPIDS and Dask, UCX-Py is an open source project under development. Akshay Venkatesh, one of the UCX-Py developers, presented a good introduction to UCX-Py at Nvidia GPU Technology Conference (GTC) 2019 (or you can view the slides online), where he presented some initial performance results comparing UCX to UCX-Py. For small messages, UCX-Py has only slightly more latency than UCX, but it is virtually identical to UCX for large messages. UCX-Py is just beginning. The first release, version 0.1, was announced on Septenber 28, 2019. You can install it with Conda,
$ conda create -n ucx -c conda-forge -c jakirkham/label/ucx cudatoolkit=ucx-proc=*=gpu ucx ucx-py python=3.7
or build it from source. UCX-Py has a serious role to play in allowing RAPIDS, Dask, and other tools to scale at the network level.
Dask
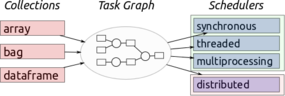
As data sets get constantly larger, at some point, they can easily become too large for a single node, resulting in the need to use multiple nodes just to hold and compute the data (parallel computing). The lead developer of Dask, Matthew Rocklin, gave a one-sentence overview of Dask: "Dask is a Python library for parallel programming that leverages task scheduling for computational problems." Dask’s initial focus was on machine learning, but it is now being applied to many areas of scientific or numerical computing. It is very Pythonic, allowing Python users to learn Dask quickly, and has been designed with three familiar interfaces: (1) array collections, (2) dataframe collections, and (3) bag collections.
One important aspect of Dask that often goes unnoticed is that the interfaces have been designed to work with "out-of-core" data – that is, data arrays that might be too large to keep in memory and "spill out" into data files. Dask arrays scale NumPy-like arrays or anything that looks like a NumPy array, such as the GPU-based CuPy, which takes NumPy code and makes it scalable across distributed systems. If you are familiar with NumPy coding, then using Dask arrays will not be a big change. This familiarity also extends to scikit-learn, which can use NumPy data types.
Dask dataframes, such as those from pandas or pandas-like structures (e.g., RAPIDS cuDF), scale, and you can use the pandas functions and methods with which you are already familiar. Dask bags are a bit different, in that they are not really designed to imitate a particular tool or data collection. Rather, they deal with collections of generic Python objects, occupying a small memory footprint and using Python iterators. You can think of it as a parallel version of PyToolz.
Dask bags allow you to use other data types or collections not covered by NumPy-like arrays or pandas-like dataframes. These high-level collections operate in parallel on datasets, even those that don't fit into main memory, and are very useful and important for large datasets in arrays, dataframes, or bags or when more performance is needed (i.e., running algorithms in parallel). As an example, you can use common pandas operations (e.g., groupby, join, and time series computations) in Dask dataframes. However, these are operated on in parallel for large data sets and for situations requiring higher performance.
Another example is for Dask bags, which are commonly used to parallelize simple computations on unstructured or semi-structured data, like text data, logfiles, JSON records, or user-defined Python objects. Operators such as map, filter, fold, and groupby can be used.
To better understand Dask, look at the simple example in Listing 1 that illustrates the close relationship between pandas and Dask.
Listing 1: Dask and pandas Similarities
| pandas | Dask |
import pandas as pd
df = pd.read_csv('2015-01-01.csv')
df.groupby(df.user_id).vaue.mean()
|
import dask.dataframe as dd
df = add.read_csv('2015-*-*.csv')
df.groupby(df.user_id).value.mean()
|
Porting code between the two is very easy. Dask is designed to run very fast and efficiently, especially on Dask arrays, Dask dataframes, and the following operations:
- Arithmetic
- Common aggregations
- Calls to apply
- Calls to value_counts() and drop_duplicates() or corr()
- Filtering with loc, isin, and row-wise selections
Beyond data collection, a second very important capability is the dynamic task scheduler that runs Python task graphs on distributed systems such as clusters. Dask allows custom, user-defined workloads, as well. During execution of the graph, Dask workers can be added or subtracted as needed, which is in contrast to other data messaging, in which the number of workers is, for the most part, fixed (e.g., with MPI, although it does have dynamic aspects). Losing an MPI rank during execution can be disastrous, whereas Dask can just continue using another worker and continue computing.
The schedulers can be broken into a few general categories: (1) synchronous, (2) threaded, (3) multiprocessing, and (4) distributed. These schedulers are designed to run on everything from a laptop for personal use, to thousand-node clusters. To run, Dask takes large-scale data collections and creates a task graph (Figure 1).
Each node of the task graph is a normal Python function, and the graph edges between nodes are normal Python objects created by one task as outputs and used by another task as inputs. Once these graphs are created, they need to be executed on parallel hardware. The task scheduler takes care of execution with of one of the previously mentioned schedulers.
The task scheduler(s) and execution graphs constitute a powerful workflow. For example, if a worker suddenly stops, the scheduler can just start using another worker, and no processing is lost. Also, if needed, additional workers can be used either for performance reasons or to provide more computational power if the graph needs it at some point.
Machines like laptops use a "single machine scheduler" that provides basic features on a local process or thread pool. It is simple and cheap to use for a single node because it doesn't have to scale. Don't forget that Dask can use out-of-core datasets, so you don't have to fit everything into memory. The performance may be a bit slower, but you can still run the workflows on your laptop.
Distributed resources use distributed schedulers, which are more sophisticated and require a requisite amount of effort to configure. The resulting workflow can then be run across distributed resources such as a cluster. To repeat: One of the really cool things about Dask is that it can expand to use additional resources (nodes) as needed to process the graph, or it can stop using resources if they aren't needed. This is a bit different from classic HPC distributed systems that usually use a fixed number of resources.
Dynamic resource usage doesn't usually fit into the HPC world because the sharing of resources can cause problems, such as the overuse of memory or the use of too many compute resources (cores); however, dask-mpi lets you use Dask in the HPC world from within an existing MPI environment. Dask takes some work and practice to learn. An online tutorial walks you through a simple Dask example with various options and talks about the effect on overall performance. A large number of tutorials and examples can be found around the web, so take a look at them to get started. The next few sections discuss the integration of Dask and … well, everything!
Dask and UCX
The recent UCX-Py (previously discussed) integration with Dask allows Python code executed via Dask to utilize UCX. If you have a fast network (e.g., InfiniBand), the Dask tasks that require coordination and communication are minimized as much as possible. A couple of fairly recent articles on high-performance networking with UCX and DGX and Python and GPUs briefly discuss the integration of UCX, via UCX-Py, and Dask.
Dask + RAPIDS
Pandas, scikit-learn, and NumPy were not designed to scale; they were designed for single-image systems (a single node). Dask, along with other tools, was designed to scale to multiple machines. Dask integrated with UCX results in a scalable set of open source Python-based tools for better performance and the use of large datasets. The obvious next step was to integrate all of this with RAPIDS, an open source set of tools for running Machine Learning (ML) applications and general numerical Python applications on GPUs. RAPIDS focuses on GPU-compatible versions of pandas, scikit-learn, and NumPy (the GPU version called CuPy).
Dask + MPI
Dask is a very cool tool for running Python code – primarily pandas, scikit-learn, and NumPy – with large data sets in parallel. It creates a task graph of how the computations take place and then executes those tasks on the available resources. It might add or subtract nodes during execution to match the tasks in the graph or to improve performance. This innovative approach, however, does not really work well in current HPC environments. Although you can restrict Dask to use a fixed number of nodes, integrating the node names and capabilities can be problematic. To better integrate Dask into HPC environments, Dask-MPI was created so you could deploy Dask applications easily within an existing MPI environment using MPI command-line launchers such as mpirun and mpiexec. It provides a convenient interface for launching your Dask cluster from within a classic HPC batch script or directly from the command line. For example, you can launch a Dask cluster from the command line with dask-mpi, specifying a "scheduler JSON" file:
$ mpirun -np 4 dask-mpi --scheduler-file /path/to/scheduler.json
Then, you can access the cluster from a batch script or an interactive session (e.g., a Jupyter Notebook) by referencing the scheduler file:
from dask.distributed import Client client = Client(scheduler_file='/path/to/scheduler.json')
Alternatively, you can turn the batch Python script into an MPI executable with the initialize function,
from dask_mpi import initialize initialize() from dask.distributed import Client client = Client() # Connect this local process to remote workers
and launch your Python script directly with miprun or mpiexec:
$ mpirun -np 4 python my_client_script.py
Take a look at the latest version of the Dask-MPI documents or the dask-mpi GitHub site to learn more. Dask-MPI is a great tool for integrating Dask applications into HPC environments.
Interoperability
All of the tools mentioned in this Python series are focused on making Python faster and more scalable. Although interoperation on data in memory on CPUs was straightforward, interoperability with GPU-based tools was not so good. It was impossible to get the tool to use the data in GPU memory without bringing the data back to the CPU and resending the data to the GPU in the form another tool could use, needlessly reducing performance because of data movement over the slow PCIe bus (although some CPUs communicate with GPUs over NVLink, which has much better performance than the PCIe). The rise of the GOAI initiative and the development of cuDF allowed a whole range of Python GPU tools to interoperate. Finally, you have one tool that uses cuDF and leaves the data on the GPU for a different tool to take over.
This article presents options for writing distributed Python applications. Mpi4py provides the familiar MPI APIs for data passing across nodes. Dask provides a Pythonic environment – primarily for pandas, scikit-learn, and NumPy applications – for distributed systems. Tracking the various Python tools, you now have a scalable ecosystem of pandas, scikit-learn, and NumPy, with GPU versions of these tools courtesy of RAPIDS and a way to write MPI code with them (mpi4py). The Pythonic Dask tool lets you run workflows with distributed versions of these tools and works with out-of-core data sets, as well. A very high performance communication framework is also incorporated to improve scalability and performance. All of these tools can be used standalone, or they can be used together in a larger ecosystem of tools. The point is, they all interoperate.
Summary
This is the final article in the Python series. Python, an amazing language with a huge number of modules is starting to dominate various technical and non-technical areas as the language of choice. I hope these articles have presented some of the more common tools for improving the performance of numerically intense computations. The tools allow you to write code in C/C++ or Fortran and call it from within the Python interpreter. You can also use some of these tools to run OpenMP code that lets you use all available CPU cores. Python code can also be compiled with a just-in-time (JIT) compiler to improve performance by using all of the CPU cores and by running Python code on GPUs. Other tools allow you to run code on GPUs from within Python. Finally are the tools that allow Python to run on distributed environments on top of very high performance network frameworks and the Pythonic tools for running code built with NumPy, pandas, and scikit-learn, although you can also run general Python code. I hope you appreciate the rich variety of tools and how they interoperate.