In the last of three articles on HDF5, we explore performing parallel I/O with the use of HDF5 files.
Parallel I/O for HPC
Amdahl's law says that your application will only go as fast as its serial portion. As the application is run over more processors, the decrease in run time gets smaller. As the number of processors, N, goes to infinity, the wall clock time approaches a constant. In general, this constant can be thought of as the “serial time” of the application. That is, the amount of time the application needs to run regardless of the number of processes used.
Many times I/O is a significant portion of this serial time. If the I/O could be parallelized, the application could scale even further, which could improve application performance and the ability to run larger problems. The great thing about HDF5 is that, behind the scenes, it is performing MPI-IO. A great deal of time has been spent designing and tuning MPI-IO for various filesystems and applications, which has resulted in very good parallel I/O performance from MPI applications.
In this article, I explore using HDF5 for parallel I/O. The intent is to reduce the I/O portion of serial time to improve scalability and performance. Before jumping knee-deep into parallel I/O to a single dataset in an HDF5 file, I’ll explore each MPI process writing to a separate dataset in the same HDF5 file.
Single Thread–Process I/O
In a previous article, I presented several ways for applications to perform I/O. One of the approaches is either for each MPI task to write to a specific part of the file relative to all other tasks (to avoid data corruption) or for each task to write a specific set of data to the file. This second approach can be used with HDF5. The great thing about HDF5 is that all of the data can be written to the same file.
The example (Listing 1) is fairly simple and illustrates how the basics of parallel I/O work with HDF5. The example will be written in Fortran for two reasons: (1) it's easy to read and (2) it's the scientific language of kings and scientists.
Listing 1: Basic Parallel I/O with HDF5
1 PROGRAM TEST1 2 3 USE iso_c_binding 4 USE HDF5 ! This module contains all necessary modules 5 IMPLICIT NONE 6 7 INCLUDE 'mpif.h' 8 9 INTEGER :: NUMTASKS, MPI_RANK, LEN, IERR , MPIERROR, INFO, C_LEN 10 INTEGER :: I, J 11 INTEGER(HID_T) :: PLIST_ID ! Property list identifier 12 INTEGER(HID_T) :: DCPL 13 INTEGER(HID_T) :: FILE_ID ! File identifier 14 INTEGER(HID_T) :: DSET_ID ! Dataset identifier 15 INTEGER(HID_T) :: FILESPACE ! Dataspace identifier in file 16 INTEGER(HSIZE_T), DIMENSION(2) :: DIMSF = (/5,8/) ! Dataset dimensions 17 INTEGER, ALLOCATABLE, TARGET :: DATA(:,:) ! Data to write 18 INTEGER :: RANK = 2 ! Dataset rank 19 20 CHARACTER(MPI_MAX_PROCESSOR_NAME) HOSTNAME 21 CHARACTER(LEN=100) :: FILENAME ! File name 22 CHARACTER(LEN=3) :: C ! Dataset name for specific rank 23 CHARACTER(LEN=10) :: DATASET_NAME 24 25 TYPE(C_PTR) :: F_PTR 26 ! ---------------------------------------------------------------------- 27 ! Initialization 28 INFO = MPI_INFO_NULL 29 30 FILENAME = "test1.hdf5" ! Define File name 31 32 ! Initialize MPI 33 CALL MPI_INIT(IERR) 34 35 ! Get number of tasks 36 CALL MPI_COMM_SIZE(MPI_COMM_WORLD, NUMTASKS, MPIERROR) 37 38 ! Get my rank 39 CALL MPI_COMM_RANK(MPI_COMM_WORLD, MPI_RANK, MPIERROR) 40 41 ! Initialize FORTRAN interface 42 CALL H5OPEN_F(IERR) 43 44 ! Initialize data buffer with trivial data 45 ALLOCATE ( DATA(DIMSF(1),DIMSF(2)) ) 46 DO I = 1, DIMSF(2) 47 DO J = 1, DIMSF(1) 48 DATA(J,I) = J - 1 + (I-1)*DIMSF(1)*(MPI_RANK+1) 49 ENDDO 50 ENDDO 51 52 ! Have the rank 0 process create the HDF5 data layout 53 IF (MPI_RANK == 0) THEN 54 55 ! Create the HDF5 file 56 CALL h5fcreate_f(FILENAME, H5F_ACC_TRUNC_F, FILE_ID, IERR) 57 58 ! Create the data space for the dataset 59 CALL h5screate_simple_f(RANK, DIMSF, FILESPACE, IERR) 60 61 ! Create the properities for the data 62 CALL H5Pcreate_f(H5P_DATASET_CREATE_F, DCPL, IERR) 63 64 ! This is required for this data pattern 65 CALL H5Pset_alloc_time_f(DCPL, H5D_ALLOC_TIME_EARLY_F, IERR) 66 67 ! Create each dataset with default properties 68 DO I=1,NUMTASKS 69 WRITE(C,"(i0)") i 70 dataset_name = "dataset" // TRIM(C) 71 WRITE(*,*) 'C = ',C,' data_name = ', DATASET_NAME 72 CALL h5dcreate_f(FILE_ID, DATASET_NAME, H5T_NATIVE_INTEGER, & 73 FILESPACE, DSET_ID, IERR, DCPL_ID=DCPL) 74 CALL h5dclose_f(DSET_ID, IERR) 75 END DO 76 77 ! Close the data space 78 CALL h5sclose_f(FILESPACE, IERR) 79 80 ! Close the properties 81 CALL h5pclose_f(DCPL, IERR) 82 83 ! Close the file 84 CALL h5fclose_f(FILE_ID, IERR) 85 END IF 86 87 ! Use an MPI barrier to make sure every thing is synched 88 CALL mpi_barrier(MPI_COMM_WORLD, IERR) 89 90 ! Setup file access property list with parallel i/o access. 91 CALL h5pcreate_f(H5P_FILE_ACCESS_F, PLIST_ID, IERR) 92 CALL h5pset_fapl_mpio_f(PLIST_ID, MPI_COMM_WORLD, INFO, IERR) 93 94 ! Open HDF5 to write data 95 CALL h5fopen_f(FILENAME, H5F_ACC_RDWR_F, FILE_ID, IERR, PLIST_ID) 96 97 ! Close the property list 98 CALL h5pclose_f(PLIST_ID, IERR) 99 100 ! Create the dataset names based on MPI rank 101 WRITE(C,"(i0)") MPI_RANK + 1 102 DATASET_NAME = "dataset" // TRIM(C) 103 104 ! Open dataset (each rank opens its own dataset) 105 CALL h5dopen_f(FILE_ID, DATASET_NAME, DSET_ID, IERR) 106 107 ! Open properties and definee MPIO model (independent) 108 CALL h5pcreate_f(H5P_DATASET_XFER_F, PLIST_ID, IERR) 109 CALL h5pset_dxpl_mpio_f(PLIST_ID, H5FD_MPIO_INDEPENDENT_F, IERR) 110 111 ! Write data to dataset (use Fortran pointer) 112 F_PTR = c_loc(DATA(1,1)) 113 CALL h5dwrite_f(DSET_ID, H5T_NATIVE_INTEGER, F_PTR, IERR) 114 115 ! Close the property list, the data set and the file 116 CALL h5pclose_f(PLIST_ID, IERR) 117 CALL h5dclose_f(DSET_ID,IERR) 118 CALL h5fclose_f(FILE_ID, IERR) 119 120 ! Close fortran interface 121 CALL h5close_f(IERR) 122 123 ! Deallocate Fortran data 124 DEALLOCATE(DATA) 125 126 ! Finalize MPI 127 CALL mpi_finalize(IERR) 128 129 END
The basic approach is for each MPI process to write an individual dataset to the same HDF5 file. To do this, the rank 0 MPI process initializes the HDF5 file and the data space with the appropriate properties and then closes the file. After that, each MPI process reopens the file, writes its data to the HDF5 file, and closes the HDF5 file. For the sake of brevity, the error codes that are returned from functions are not checked.
The initialization of the HDF5 file and properties is done by the rank 0 process starting on line 53. Rather than walk through the code explaining each function call, the following lists describe the various stages the code goes through and the corresponding lines in the code.
- The following tasks are performed by the rank 0 process, which defines the HDF5 file and its attributes.
- Line 56, create HDF5 file
- Line 59, create data space
- Line 62, start properties of file
- Line 65, call required when each process writes to a single file
- Lines 68–75, loop over all processes, create dataset name, create dataset for each process
- Line 78, close the data space
- Line 81, close properties list
- Line 84, close HDF5 file
- The following tasks are then performed by all of the processes:
- Line 91, create new property list
- Line 92, set MPI-IO property
- Line 95, open file
- Line 98, close properties
- Line 105, open dataset
- Line 109, set MPI-IO property
- Lines 112–113, create pointer to data and write data to dataset (specific dataset for each process)
- Lines 116–127, close everything and finish up
A quick run of the example in Listing 1 with two processes creates the file test1.hdf5. Listing 2 shows the content of this file using the HDF5 tool h5dump.
Listing 2: Content of HDF5 File
[laytonjb@laytonjb-Lenovo-G50-45 CODE]$ h5dump test1.hdf5
HDF5 "test1.hdf5" {
GROUP "/" {
DATASET "dataset1" {
DATATYPE H5T_STD_I32LE
DATASPACE SIMPLE { ( 8, 5 ) / ( 8, 5 ) }
DATA {
(0,0): 0, 1, 2, 3, 4,
(1,0): 5, 6, 7, 8, 9,
(2,0): 10, 11, 12, 13, 14,
(3,0): 15, 16, 17, 18, 19,
(4,0): 20, 21, 22, 23, 24,
(5,0): 25, 26, 27, 28, 29,
(6,0): 30, 31, 32, 33, 34,
(7,0): 35, 36, 37, 38, 39
}
}
DATASET "dataset2" {
DATATYPE H5T_STD_I32LE
DATASPACE SIMPLE { ( 8, 5 ) / ( 8, 5 ) }
DATA {
(0,0): 0, 1, 2, 3, 4,
(1,0): 10, 11, 12, 13, 14,
(2,0): 20, 21, 22, 23, 24,
(3,0): 30, 31, 32, 33, 34,
(4,0): 40, 41, 42, 43, 44,
(5,0): 50, 51, 52, 53, 54,
(6,0): 60, 61, 62, 63, 64,
(7,0): 70, 71, 72, 73, 74
}
}
}
}Both datasets are in the file, so each MPI process wrote its respective dataset.
Parallel I/O and HDF5
The previous discussion demonstrates a good way to get started with parallel I/O and HDF5, but it has some limitations. For example, each MPI process writes its portion of an array to a different dataset in the HDF5 file, so if you want to restart the application from the beginning, you would have to use the same number of MPI processes as were originally used. A better solution would be for each MPI process to read and write its data to the same dataset, which allows the number of MPI processes to change without having to worry about how the data is written in the HDF5 file.
The best way to achieve this is using hyperslabs. In HDF, a hyperslab is a portion of a dataset. It can be a contiguous section of a dataset, such as a block, a regular pattern of individual data values, or a block within a dataset (Table 1).
Table 1: This pattern of 2x2 blocks each are separated by a column and a row. Each 2x2 block can be a hyperslab, or each row or column of a 2x2 block can be a hyperslab. You have a number of options depending on the design.
| X | X | X | X | ||
| X | X | X | X | ||
| X | X | X | X | ||
| X | X | X | X | ||
To completely describe a hyperslab, four parameters are needed:
- start – A starting location for the hyperslab
- stride – The number of elements that separate each element or block to be selected
- count – The number of elements or blocks to select along each dimension
- block – The size of the block selected from the data space
Each of the parameters is an array with a rank that is the same as the data space.
The HDF group has created several parallel examples. The simplest is ph5example, which illustrates how to get started. In the code, the HDF5 calls are accomplished with MPI processes – which all have the same data – including writing the data to the data space. This example is a great way to state parallel I/O. In fact, the example in Listing 1, in which each MPI process wrote its own dataset, started with this example.
HDF5 has a number of examples that use hyperslab concepts for parallel I/O. Each MPI process writes a part of the data to the common dataset. The main page for these tutorials has four hyperslab examples:
- Writing datasets by contiguous hyperslab
- Writing to a dataset by regularly spaced data
- Writing to a dataset by pattern
- Writing to a dataset by chunk
In this article, I go through the last example, which writes data to a common dataset by chunks.
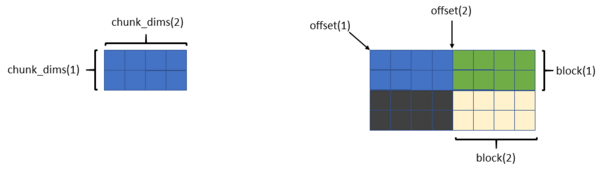
In the Fortran example the number of processes is fixed at four to illustrate how each MPI process writes to a common dataset (Figure 1). The dataset is 4x8 (four rows and eight columns), and each chunk is 2x4 (two rows and four columns).
Recall that four parameters describe a hyperslab: start, stride, count, and block. The start parameter is also referred to as offset in the example (and other) code.
The dimensions of each chunk are given by the arraychunk_dims. The first element is the width of the chunk (number of columns) and the second element is the height of the chunk (number of rows). For all MPI processes, block(1) = chunk_dims(1) and block(2) = chunk_dims(2).
Because the data from each process is a chunk, the stride array for each chunk is 1 (stride(1) = 1, stride(2) = 1); that is, the chunk is contiguous. Some of the other examples have arrays for which the stride is not 1.
The array count for each chunk is also 1; that is, each MPI process is only writing a single chunk (count(1) = 1, and count(2) = 1).
Where things differ is the offset or start of each chunk. For the sake of clarity, the rank = 0 process writes the chunk in the bottom left portion of the dataset. Therefore, both elements of its offset arrays are 0. The rank = 1 process writes the bottom right chunk of the dataset. The offset array for it is offset(1) = chunk_dims(1) and offset(2) = 0.
MPI process 2 writes the toplefthand chunk. Its offset array is offset(1) = 0 and offset(2) = chunk_dims(2). Finally, MPI process 3 writes the top righthand chunk. Its offset array is offset(1) = chunk_dims(1) and offset(2) = chunk_dims(2).
For each chunk, the data is an array of integers that correspond to (rank of the MPI process)+1. The data in the rank 0 process has values of 1, the data in the rank 1 process has values of 2, and so on.
The process for writing hyperslab data to a single dataset is a little different from the first example, but for the most part, it is the same. When creating the data space you have to call some extra functions to configure the hyperslabs.
To write the hyperslab to the dataset, each MPI process first calls h5sselect_hyperslab_f to select the appropriate hyperslab using the four parameters mentioned previously. The code then has all of the MPI processes do a collective write, which puts the information into the header of the data set. The second call writes the dataset to the location defined by the hyperslab for that process.
Running the code produces an HDF5 file named sds_chnk.h5. If h5dump is run against it, you’ll se the output shown in Listing 3.
Listing 3: Hyperslab Data
[laytonjb@laytonjb-Lenovo-G50-45 CODE]$ h5dump sds_chnk.h5
HDF5 "sds_chnk.h5" {
GROUP "/" {
DATASET "IntArray" {
DATATYPE H5T_STD_I32LE
DATASPACE SIMPLE { ( 8, 4 ) / ( 8, 4 ) }
DATA {
(0,0): 1, 1, 2, 2,
(1,0): 1, 1, 2, 2,
(2,0): 1, 1, 2, 2,
(3,0): 1, 1, 2, 2,
(4,0): 3, 3, 4, 4,
(5,0): 3, 3, 4, 4,
(6,0): 3, 3, 4, 4,
(7,0): 3, 3, 4, 4
}
}
}
}Because h5dump is written in C, the data is written to stdout in row-major C format (which is the opposite column-major Fortran). With some transposing, you can see that the dataset is exactly as expected.
It takes some work to write hyperslabs to the same dataset using MPI-IO. Read through the other examples, particularly the writing to a dataset by pattern option, to understand how hyperslabs and MPI-IO can be used to reduce the I/O time of your application.